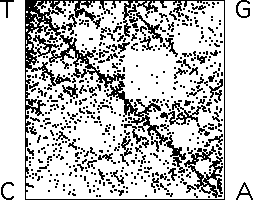
| Here is an example first
explored by H. Joel Jeffrey. The genetic code
is written in an alphabet of four characters: C, A, T, and G. A sequence
of several billion of these makes each of us. A sequence of 3957 symbols
is needed to encode the formation of the enzyme amylase. |
| How can we convert a DNA sequence into an IFS picture? |
| Read the sequence in order, and |
| apply T1 whenever C is encountered, |
| apply T2 whenever A is encountered, |
| apply T3 whenever T is encountered, and |
| apply T4 whenever G is encountered, |
|
| for example. Of course, any assignment of transformations to C, A, T, and G
can be used. |
| On the left is the picture that results from applying these rules to the
amylase sequence. Note there are very few points in the
region with address GA. (Remember the ordering of the addresses.) |
| On the
right is a picture that results when 3957 points are generated randomly,
except that T4 never immediately follows T1. |
|
| Not an exact match, but certainly suggestive.
Here are a few more examples. |